How to contribute annotations to PR2 reference database
Steps
- Contact one member of the Core Team
- Explain which group you want to annotate. It can be a genus, a class or any other taxonomic level.
- We will send you an Excel file and a fasta file containing all existing PR2 sequences for the group you are expert in.
- You can alternatively download the data from the web interface
- Follow the instructions below to update or add data.
- Send back the updated Excel file
- Your contribution will be added to the next release of PR2 (we are doing 2 to 3 releases per year).
- You will be acknowledged as a contributor on the PR2 web site and you name will appear as a co-author on the Zenodo entry for the next PR2 update (see for example)
Files provided
Three files will be provided to you
- A fasta file with the PR2 accession number
- A text file that can be used to annotate trees
- An excel file with 2 sheets (taxonomy, metadata)
Please edit the Excel file by marking all your changes in yellow.
pr2_export.fas.gz - FASTA file
Contains the sequence labelled with the PR2 accession number
>JX988758.1.1807_U
TTGATCCTGCCAGTAGTCATATGCTTGTCTCAAAGATTAAGCCATGCATGTCTAAGTATAAGCACCTTATACTGTGAAACTGCGAATGGCTCATTAAATCAGTTATCGTTTATTTGATGATCTCTTGCTACTTGGATACCCGTGGT...
...
pr2_export.txt
- Contains information about each sequence that can be uploaded to TreeView or TreeViewer.
- Fields are separated by tabulation.
- Species that have been assigned automatically by dada2 are labelled with 1 in the column
pr2_annotated. - You can generate more complete text files by exporting columns of the
metadatasheet from the Excel filepr2_export.xlsx.
pr2_accession domain supergroup division subdivision class order family genus species pr2_annotated gb_strain
AF265331.1.1123_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifila Amphifila_marina 1
AY082983.1.1879_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifila Amphifila_sp. 1
EF023442.1.1807_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifilaceae_X Amphifilaceae_X_sp. 1
EF023338.1.1806_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifilaceae_X Amphifilaceae_X_sp. 1
EF023208.1.1805_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifilaceae_X Amphifilaceae_X_sp. 1
EF023658.1.1802_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifilaceae_X Amphifilaceae_X_sp. 1
EF023821.1.1802_U Eukaryota TSAR Stramenopiles Bigyra Sagenista Labyrinthulomycetes Amphifilaceae Amphifilaceae_X Amphifilaceae_X_sp. 1
...
pr2_export.xlsx - Excel file
This file contains two sheets:
Taxonomy - DO NOT EDIT
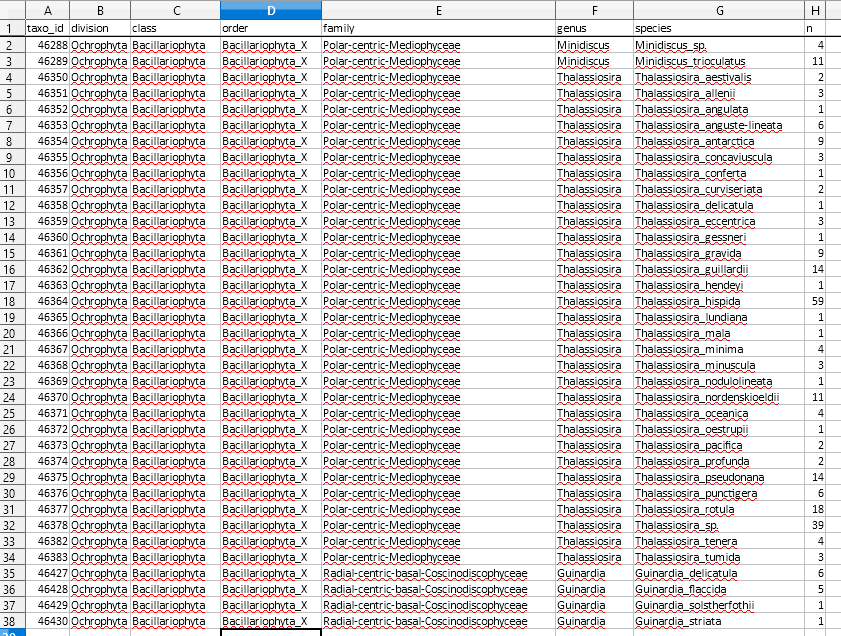
- This sheet provides a summary of the current taxonomy of the group with the number of sequences for each species (n).
- Please do not edit this file directly, this is only for your information.
Excel - Metadata - edit only this file
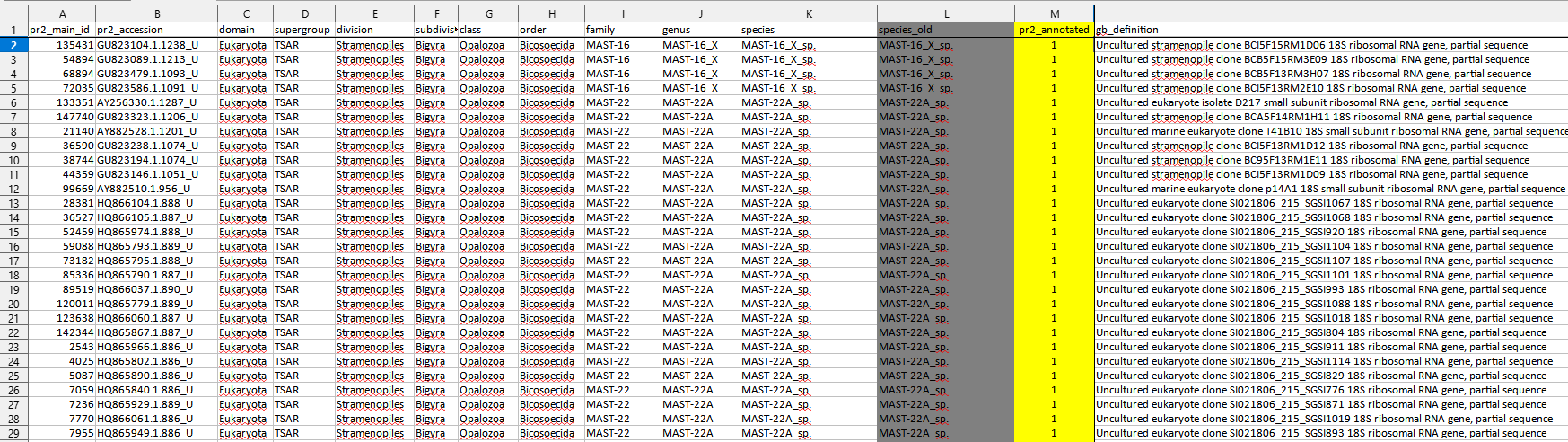
- Each sequence has a unique identifier (pr2_accession) which is based on the GenBank accession (genbank_accession).
- For each sequence, the full taxonomic path is provided along with metadata (see here for a full description of the fields).
- There are two types of entries (column
pr2_annotated):- If
pr2_annotatedis equal to 1, the entry is part of the reference PR2 database and has been previously validated - If
pr2_annotatedis equal to 0, the entry is NOT part of the reference PR2 database. It has been automatically annotated using dada2 AssignTaxonomy. It needs to be validated and you can validate it (changepr2_annotatedto 1 and mark in yellow).
- If
Modifying or adding entries
Only change entries in the Metadata table
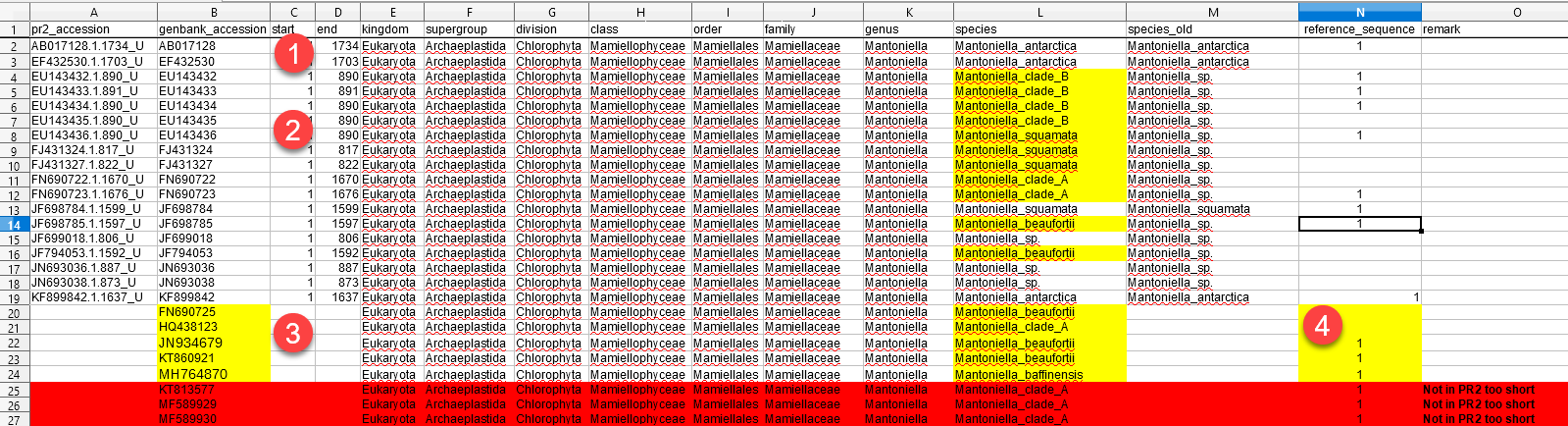
- You can
- modify the taxonomy of a given entry (do not alter the
species_oldcolumn though). - add new metadata. If your metadata do not fit the existing columns, just add more columns and we will see how to incorporate them.
- modify the taxonomy of a given entry (do not alter the
- You can change the ranks (supergroup to genus) if necessary but you must make sure that:
- you follow exactly the PR2 conventions which are detailed here (see the second paragraph). In particular, any taxonomic name can only appear in a single column (taxonomic level). Use the _X convention to distinguish different levels with the same name.
- you are consistent for all sequences belonging to the same taxon.
- You can also add new species as needed.
- Please see the figure above for some examples of changes
- 1 - These entries are unchanged
- 2 - These entries have been reassigned to a new species
- 3 - These are new entries. Provide the following information
- genbank_accession. We will download the sequence and all genbank metadata, so no need for you to do it
- taxonomy assignation. If the species is already present in the database you can just provide the species name
- if the sequence is not limited to the 18S, but also contains the ITS, please provide the coordinates on the sequence of the start and end of the 18S rRNA gene.
- 4 - You can also indicate whether the new sequence is a reference sequence. Reference sequences are have a high quality, preferentially full length and are representative a given taxon.