Reference database structure
From version 4.14.0, a single SSU database is provided which contains sequences for:
- 18S rRNA from nuclear and nucleomorph
- 16S rRNA from plastid, apicoplast, chromatophore, mitochondrion
- 16S rRNA from a small selection of bacteria
The rationale is that the database can now be used to detect bacterial sequences that are amplified with either 18S rRNA or “universal” primers. These sequences can be further assigned with Silva or GTDB.
Taxonomy structure
Version 5.0 and above
- 9 levels : Domain / Supergroup / Division / Subdivision / Class / Order / Family / Genus / Species
Kingdomis replaced byDomain- Adding a
Subdivisionlevel
Version 4.14.1 and below
- 8 levels : Kingdom / Supergroup / Division / Class / Order / Family / Genus / Species
Rules for naming taxonomic levels
- All taxonomy paths are unique. For example for a given Class, all higher levels are similar (Division -> Supergroup -> Domain)
- No taxon can appear at different levels. For example “Stramenopiles” cannot appear at both the Supergroup and Division levels.
- No space (use underscore). For example Home_sapiens
- If species is not known use “Genus_sp.” (note the . after sp). The Genus level can be a level with Xs (see next rule): e.g. “Stramenopiles_XXXX_sp.“,
- If level i is unknown
- if level i-1 is known, use level i-1 followed by “_X” , e.g. “Stramenopiles_X” at Division level
- if level i-1 is unknown, use level i-1 followed by “X” , e.g. “Stramenopiles_XX” at Class level
Organelles
In order to allow correct assignation with software such as DECIPHER (IDTax) for organelle, the taxonomy is appended with 4 letters corresponding to the organelle
| Organelle | Taxonomy suffix |
|---|---|
| nucleus | |
| nucleomorph | :nucl |
| plastid | :plas |
| apicoplast | :apic |
| chromatophore | :chrom |
| mitochondrion | :mito |
e.g. a sequence annotated as Neoceratium_horridum:plas will correspond to a plastid sequence of the species Neoceratium horridum
Rules for sequences
- Minimum length = 500 bp (except for some plastid sequences which are shorter)
- Maximum number of ambiguities (N) = 20
- No consecutive Ns (so sequences such as ..ATGNNAT.. are removed)
Construction of PR2 id (pr2_accession)
Accession.Start.End_X
For example AF530536.1.1695_U
- AF530536 : GenBank accession number
- 1 : Start of the 18S rRNA in the GenBank sequence
- 1695 : End of the 18S rRNA in the GenBank sequence
- X can take any of the following values:
- G: genomic sequence containing a described intron (rDNA)
- R: the previous genomic rRNA sequence, without the intron(s)
- U: no intron described, but intron(s) may be present
- UC: introns were detected in silico and removed from the sequence (putative rRNA)
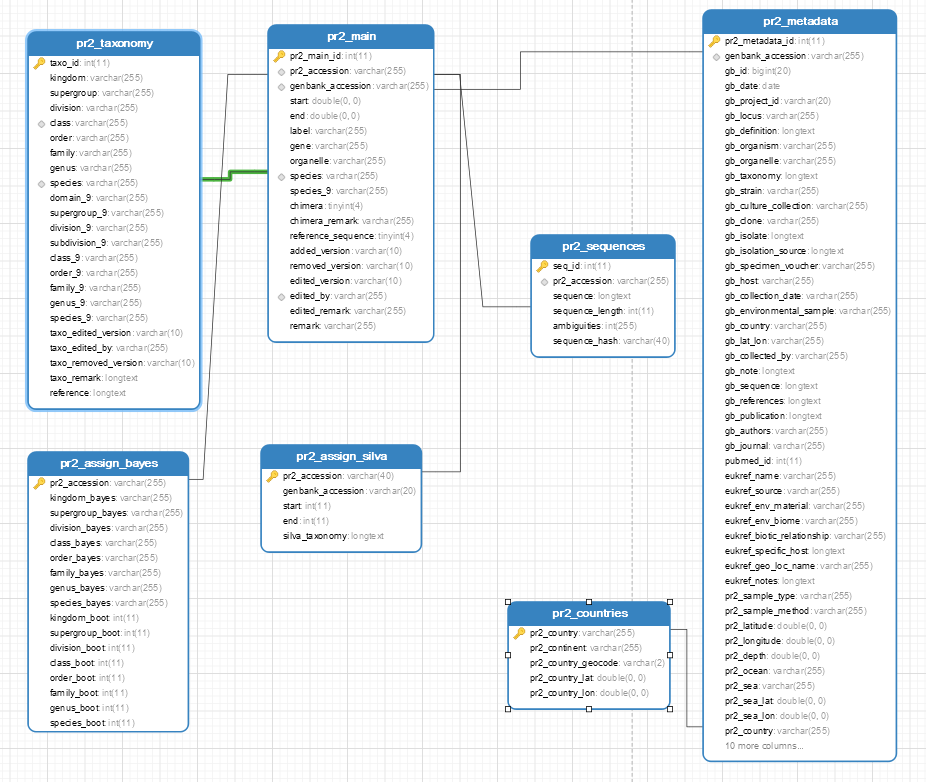
Database structure (tables and fields)
| Table | Fields | Comment |
|---|---|---|
| pr2_main | pr2_accession, genbank_accession, species, editing hitory | |
| pr2_sequence | pr2_accession, , sequence | linked to pr2_main by pr2_accession |
| pr2_metadata | metadata from GenBank and from published references | linked to pr2_main by genbank_accession |
| pr2_taxonomy | domain -> species, editing history | linked to pr2_main by species |
| pr2_countries | geo-information | linked to pr2_metadata by pr2_country |
| pr2_assign_silva | assignment of pr2 sequences according to Silva | linked to pr2_main by pr2_accession |
| eukribo_V2 | Eukribo version 2 data | linked to pr2_main by genbank_accession |
| pr2_traits | Traits from other databases harboring species traits | linked to pr2_taxonomy by taxonomy level (e.g. species, genus). This table used a long format. |
| worms_taxonomy | Worms database entries for taxa from PR2 |
History of database structure changes
| PR2 version | Table | Field | Action | Comment |
|---|---|---|---|---|
| 5.0.0 | eukribo_V2 | A | New table holding Eukribo version 2 data. | |
| pr2_traits | A | New table for adding traits to PR2 from other databases (e.g. Mixoplankton Database) | ||
| worms_taxonomy | A | New table with Worms database entries for taxa from PR2 | ||
| 4.14.0 | pr2_main | quarantined_version | A | quarantined sequences do not appear in the released version but will be re-assigned latter |
| pr2_metadata | gb_references | R | empty | |
| gb_locus | R | empty | ||
| gb_division | A | 3-letter code (e.g. PLN, ENV) | ||
| 4.13.0 | pr2_metadata | pr2_depth | A | depth of sample in meter |
| gb_id | A | Genbank ID number (big integer) | ||
| gb_project_id | A | Genbank project ID for metagenomes | ||
| gb_sequence | A | original gb_sequence (longtext) | ||
| pr2_countries | A | New table for country information | ||
| pr2_assign_silva | A | New table for Silva assignation of sequences | ||
| 4.12.0 | pr2_main | gene | A | 18S_RNA, 16S_RNA |
| organelle | A | nucleus, plastid, mitochondria, nucleomorph, apicoplast (left empty for cyanobacteria) | ||
| pr2_metadata | gb_organelle | A | import the corresponding gb field | |
| pr2_sequence_origin | M | add other possibilities such as genome and metagenome | ||
| pr2_continent, pr2_country, pr2_country_lat, pr2_country_lon | A | geographical origin extracted from gb_country field, gb_isolation_source, eukref_geo_loc_name | ||
| pr2_location, pr2_location_lat, pr2_location_lon | A | geographical origin extracted from gb_country field, gb_isolation_source, eukref_geo_loc_name | ||
| pr2_sea, pr2_sea_lat, pr2_sea_lon | A | geographical origin extracted from gb_country field, gb_isolation_source, eukref_geo_loc_name | ||
| pr2_sequence | sequence_hash | A | hash value of the sequence (using R function digest::sha1 | |
| pr2_taxonomy | taxon_trophic_mode | A | detailed trophic mode (e.g. “C-fixation constitutive; Mixotroph”) - for future use | |
| 4.11.1 | pr2_metadata | gb_reference | A | Contains the whole REFERENCE field from GenBank |
| eukref_publication eukref_authors eukref_journal | M | Merged into gb_publication, gb_authors, gb_journal | ||
| 4.11.0 | pr2_metadata | eukref_publication eukref_authors eukref_journal eukref_notes | A | Added |
| 4.11.0 | pr2_metadata | pr2_xxx | M | fields annotated by Eukref renamed back to eukref_xxx |
| 4.11.0 | pr2_main | taxo_id | R | not used anymore since linked is done by species (this will be probably reinstated later on a different form using a different format for the taxonomy table). |
| 4.9 | pr2_metadata | eukref_xxx | M | renamed to pr2_xxx to reflect the fact that these fields will also be updated independantly of EukRef |
| 4.9 | pr2_main | reference_sequence | A | designate sequences that can be used as reference e.g. for alignment |
| 4.8 | pr2_main, taxo | xx_date (e.g. edited_date) | R | redundant with xx_version (e.g. edited_version) fields |
| 4.8 | pr2_sequence | sequence_length | M | Minimum length is now 500 bp. |
Action : A=Added/R=Removed/M=Modified