Format of PR2 reference flat files
List of files are provided for each release
Link to files of latest release
Each version has a 3 number code: version_x.y.z, e.g. 5.1.0
Important note : All files are in UNIX format (end of lines are indicated Line Feed-LF only). Please see at the bottom of the file how to converts the files to Windows format (end of lines are CR+LF).
Files to assign metabarcodes
dada2
- pr2_version_x.y.z_SSU_dada2.fasta.gz contains a dada2 format compatible training file
Mothur or Qiime
Two files:
- pr2_version_x.y.z_SSU_mothur.fasta.gz contains all sequences in fasta format with the accession in the description line
- pr2_version_x.y.z_SSU_mothur.tax.gz contains the taxonomy of each sequence separated from the accession number by a tabulation
- Note :Qiime only use 7 taxonomical levels by default.
USEARCH or VSEARCH
- pr2_version_x.y.z_SSU_UTAX.fasta.gz contains one fasta file with the accession number of the sequence and its full taxonomy on the description line in the UTAX format. It is suitable to use with USEARCH and VSEARCH.
DECIPHER training set
- pr2_version_x.y.z_SSU.decipher.trained.rds is a DECIPHER training set using 3 iterations and a maximum of 20 sequences per species (LearnTaxa) that can be used with the DECIPHER IDTaxa function to assign metabarcodes. Use the readRDS() function of R to load the file.
Emu
Emu is a very efficient program to assign reads to a taxonomy database. It is in particular useful for long reads such as provided by Nanopore or PacBio sequencers.
- pr2_version_x.y.z_emu.zip .The files provided starting on version 5.1.0 are those used to build the emu database which is done with the following command:
emu build-database pr2_version_5.1.0_emu.db --sequences pr2_version_5.1.0_emu.fasta --seq2tax pr2_version_5.1.0_emu_map.tsv --taxonomy-list pr2_version_5.1.0_emu_taxo.tsv
Other files
Taxonomy structure
- pr2_version_x.y.z_taxonomy.xlsx
Fasta file
- pr2_version_x.y.z_SSU_taxo_long.fasta.gz: contains accession number, the name of sequence and full taxonomy on the description line. It is suitable to build a local database for BLAST search
Full database as Excel file
- pr2_version_x.y.z_merged.xlsx: Includes sequences, taxonomy and metadata.
Sequences annotated as chimeric
- pr2_version_x.y.z_chimera.xlsx
Unassigned sequences
pr2_version_x.y.z_unassigned.xlsx
These SSU GenBank sequences are not part yet of the PR2 database. However some of them have been automatically assigned with AssignTaxa and version 4.14 of PR2 (see columns at the end of the xlsx file).
These sequences can be useful to add more sequences to a phylogenetic tree for example or annotate a specific taxonomic group.
Examples of format
mothur fasta
>AB353770.1.1740_U
ATGCTTGTCTCAAAGATTAAGCCATGCATGTCTCAGTATAAGCTTTTACATGGCGAAACTGCGAATGGCTCATTAAAACA
GTTACAGTTTATTTGAAGGTCATTTTCTACATGGATAACTGTGGTAATTCTAGAGCTAATACATGCGCCCAAACCCGACT
CCGTGGAAGGGTTGTATTTATTAGTTACAGAACCAACCCAGGTTCGCCTGGCCATTTGGTGATTCATAATAAACGAGCGA
ATTGCACAGCCTCAGCTGGCGATGTATCATTCAAGTTTCTGACCTATCAGCTTCCGACGGTAGGGTATTGGCCTACCGTG
GCAATGACGGGTAACGGAGAATTAGGGTTCGATTCCGGAGAGGGAGCCTGAGAAACGGCTACCACATCTAAGGAAGGCAG
CAGGCGCGCAAATTACCCAATCCTGACACAGGGAGGTAGTGACAAGAAATAACAATACAGGGCAACCATGTCTTGTA`
mothur taxonomy
AB353770.1.1740_U
Eukaryota;Alveolata;Dinophyta;Dinophyceae;Dinophyceae_X;Dinophyceae_XX;Peridiniopsis;Peridiniopsis_kevei;
KC672520.1.1801_U
Eukaryota;Opisthokonta;Fungi;Ascomycota;Pezizomycotina;Leotiomycetes;Leotiomycetes_X;Leotiomycetes_X_sp.;
UTAX fasta
>AB353770.1.1740_U;tax=k:Eukaryota,d:Alveolata,p:Dinophyta,c:Dinophyceae,o:Dinophyceae_X,f:Dinophyceae_XX,g:Peridiniopsis,s:Peridiniopsis_kevei
ATGCTTGTCTCAAAGATTAAGCCATGCATGTCTCAGTATAAGCTTTTACATGGCGAAACTGCGAATGGCTCATTAAAACA
GTTACAGTTTATTTGAAGGTCATTTTCTACATGGATAACTGTGGTAATTCTAGAGCTAATACATGCGCCCAAACCCGACT
CCGTGGAAGGGTTGTATTTATTAGTTACAGAACCAACCCAGGTTCGCCTGGCCATTTGGTGATTCATAATAAACGAGCGA
ATTGCACAGCCTCAGCTGGCGATGTATCATTCAAGTTTCTGACCTATCAGCTTCCGACGGTAGGGTATTGGCCTACCGT
dada2 fasta
>Eukaryota;Alveolata;Dinophyta;Dinophyceae;Dinophyceae_X;Dinophyceae_XX;Peridiniopsis;Peridiniopsis_kevei;
ATGCTTGTCTCAAAGATTAAGCCATGCATGTCTCAGTATAAGCTTTTACATGGCGAAACTGCGAATGGCTCATTAAAACA
GTTACAGTTTATTTGAAGGTCATTTTCTACATGGATAACTGTGGTAATTCTAGAGCTAATACATGCGCCCAAACCCGACT
fasta long
>AB353770.1.1740_U||Eukaryota|Alveolata|Dinophyta|Dinophyceae|Dinophyceae_X|Dinophyceae_XX|Peridiniopsis|Peridiniopsis_kevei
ATGCTTGTCTCAAAGATTAAGCCATGCATGTCTCAGTATAAGCTTTTACATGGCGAAACTGCGAATGGCTCATTAAAACA
GTTACAGTTTATTTGAAGGTCATTTTCTACATGGATAACTGTGGTAATTCTAGAGCTAATACATGCGCCCAAACCCGACT
CCGTGGAAGGGTTGTATTTATTAGTTACAGAACCAACCCAGGTTCGCCTGGCCATTTGGTGATTCATAATAAACGAGCGA
merged
pr2_main_id pr2_accession genbank_accession start end label taxo_id.x species chimera chimera_remark added_date added_version removed_date removed_version edited_date edited_version edited_by edited_remark remark taxo_id.y kingdom supergroup division class order family genus taxo_edited_date taxo_edited_version taxo_removed_date taxo_removed_version taxo_remark reference seq_id sequence sequence_length ambiguities pr2_metadata_id gb_date gb_locus gb_definition gb_organism gb_strain gb_culture_collection gb_clone gb_isolate gb_isolation_source gb_specimen_voucher gb_host gb_collection_date gb_environmental_sample gb_country gb_lat_lon gb_collected_by gb_note pubmed_id gb_publication gb_authors gb_journal eukref_name eukref_env_material eukref_env_biome eukref_biotic_relationship eukref_specific_host eukref_geo_loc_name pr2_sample_type p2_sample_method pr2_ocean pr2_latitude pr2_longitude pr2_sequence_origin pr2_size_fraction pr2_size_fraction_min pr2_size_fraction_max metadata_remark metadata_select sequence_label
AB353770.1.1740_U AB353770 1 1740 U 2014 Peridiniopsis_kevei 2014 Eukaryota Alveolata Dinophyta Dinophyceae Dinophyceae_X Dinophyceae_XX Peridiniopsis ATGCTTGTCTCAAAGATTAAGCCATGCATGTCTCAGTATAAGCTTTTACATGGCGAAACTGCGAATGGCTCATTAAAACAGTTACAGTTTATTTGAAGGTCATTTTCTACATGGATAACTGTGGTAATTCTAGAGCTAATACATGCGCCCAAACCCGACTCCGTGGAAGGGTTGTATTTATTAGTTACAGAACCAACCCAGGTTCGCCTGGCCATTTGGTGATTCATAATAAACGAGCGAATTGCACAGCCTCAGCTGGCGATGTATCATTCAAGTTTCTGACCTATCAGCTTCCGACGGTAGGGTATTGGCCTACCGTGGCAATGACGGGTAACGGAGAATTAGGGTTCGATTCCGGAGAGGGAGCCTGAGAAACGGCTACCACATCTAAGGAAGGCAGCAGGCGCGCAAATTACCCAATCCTGACACAGGGAGGTAGTGACAAGAAATAACAATACAGGGCAACCATGTCTTGTAATTGGAATGAGTAGAATTTAAATCCCTTTACGAGTATCCATTGGAGGGCAAGTCTGGTGCCAGCAGCCGCGGTAATTCCAGCTCCAATAGCGTATATTAAAGTTGTTGCGGTTAAAAAGCTCGTAGTTGGATTTCTGTCGAAGGAGACCGGTCCGCCCTCTGGGTGAGTATCTGGATCTCTTTGGACATCTTCTTGGGGAACGTATCTGCACTTCATTGTGCGGTGCGGTACTCAAGACTTTTACTTTGAGGAAATTAGAGTGTTTCAAGCAGGCACACGCCTTGAATACATTAGCATGGAATAATAAGATAGGACCTCGGTTCTATTTTGTTGGTTTCTAGAGCTGAGGTAATGATTAATAGGGATAGTTGGGGGCATTCGTATTTAACTGTCAGAGGTGAAATTCTTGGATTTGTTAAAGACGGACTACTGCGAAAGCATTTGCCAAGGATGTTTTCATTGATCAAGAACGAAAGTTAGGGGATCGAAGACGATCAGATACCGTCGTAGTCTTAACCATAAACCATGCCGACTAGAGATTGGAGGTCGTTATCCGTACGACTCCTTCAGCACCTTATGAGAAATCAAAGTCTTTGGGTTCCGGGGGGAGTATGGTCGCAAGGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACCAGGAGTGGAGCCTGCGGCTTAATTTGACTCAACACGGGGAAACTTACCAGGTCCAGACATAGTAAGGATTGACAGATTGATAGCTCTTTCTTGATTCTATGGGTGGTGGTGCATGGCCGTTCTTAGTTGGTGGAGTGATTTGTCTGGTTAATTCCGTTAACGAACGAGACCTTAACCTGCTAAATAGTGACACATTACCCCGGTAATGTGGGTTACTTCTTAGAGGGACTTTGCGTGTCTAACGCAAGGAAGTTTGAGGCAATAACAGGTCTGTGATGCCCTTAGATGTTCTGGGCTGCACGCGCGCTACACTGATGCGCTCAACGAGTTTATGACCTTGCCCGGAAGGGTTGGGTAATCTTGTTAAAACGCATCGTGATGGGGATAGATTATTGCAATTATTAATCTTCAACGAGGAATTCCTAGTAAGCGCGAGTCATCAGCTCGTGCTGATTAAGTCCCTGCCCTTTGTACACACCGCCCGTCGCTCCTACCGATTGAGTGATCCGGTGAATAATTCGGACCGCAGCATTTGTCAGTTCCTGACTCATGCCGTGGAAAGTCTAGTGAACCTTATCACTTAGAGGAAGGAGAAGTCGTAACA 1740 0 3830 PLN Peridiniopsis cf. kevei gene for 18S rRNA, partial sequence. 26-juil.-03 Japan: Toyama, Tomi-iwa Canal Park
Converting files from UNIX to Windows format
All files are in UNIX format (end of lines are indicated Line Feed-LF only). File can be converted to Windows format (end of lines are CR+LF):
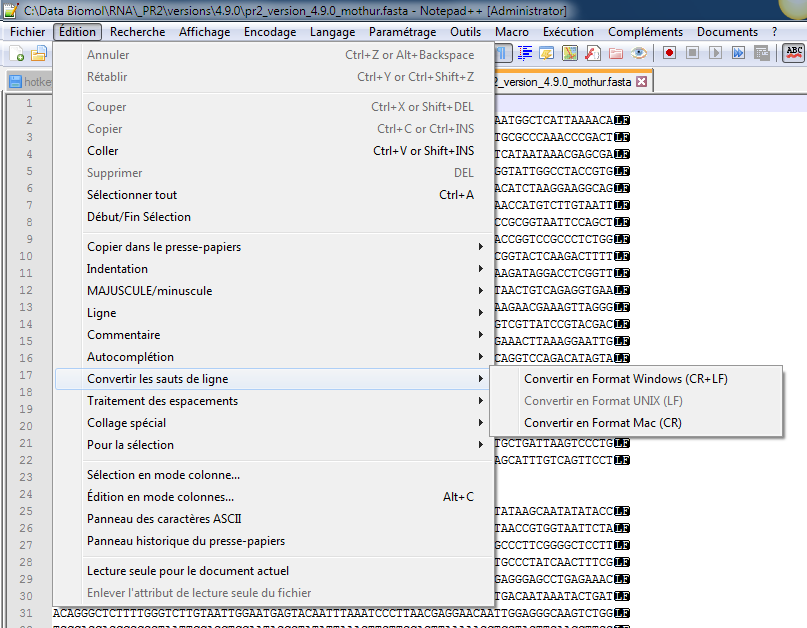
Install Notepad++
Open the files to convert (e.g. for mothur the
.fastaand.taxfiles)
- Convert the end of lines
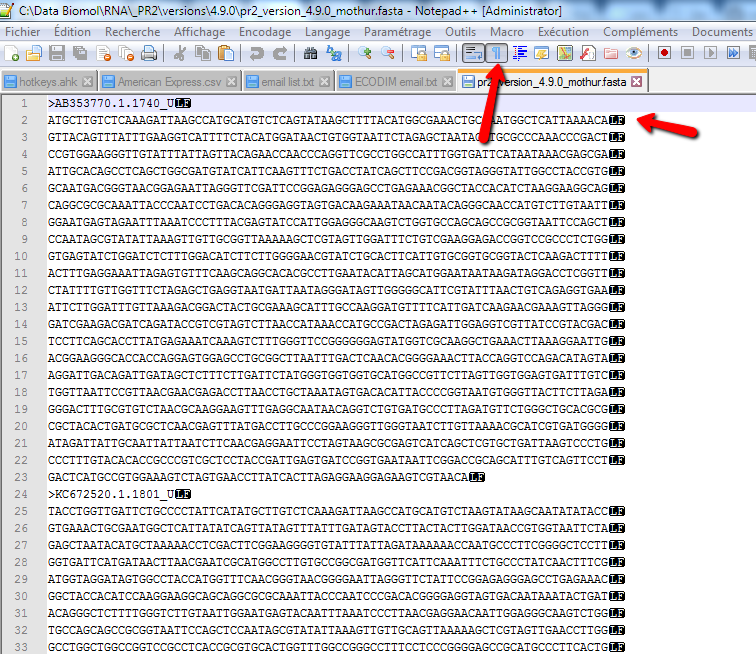
- Now the file should like the image below (red arrow)
- Save the file under the same name by clicking on the disk icon (blue arrow)