The 9-levels taxonomy
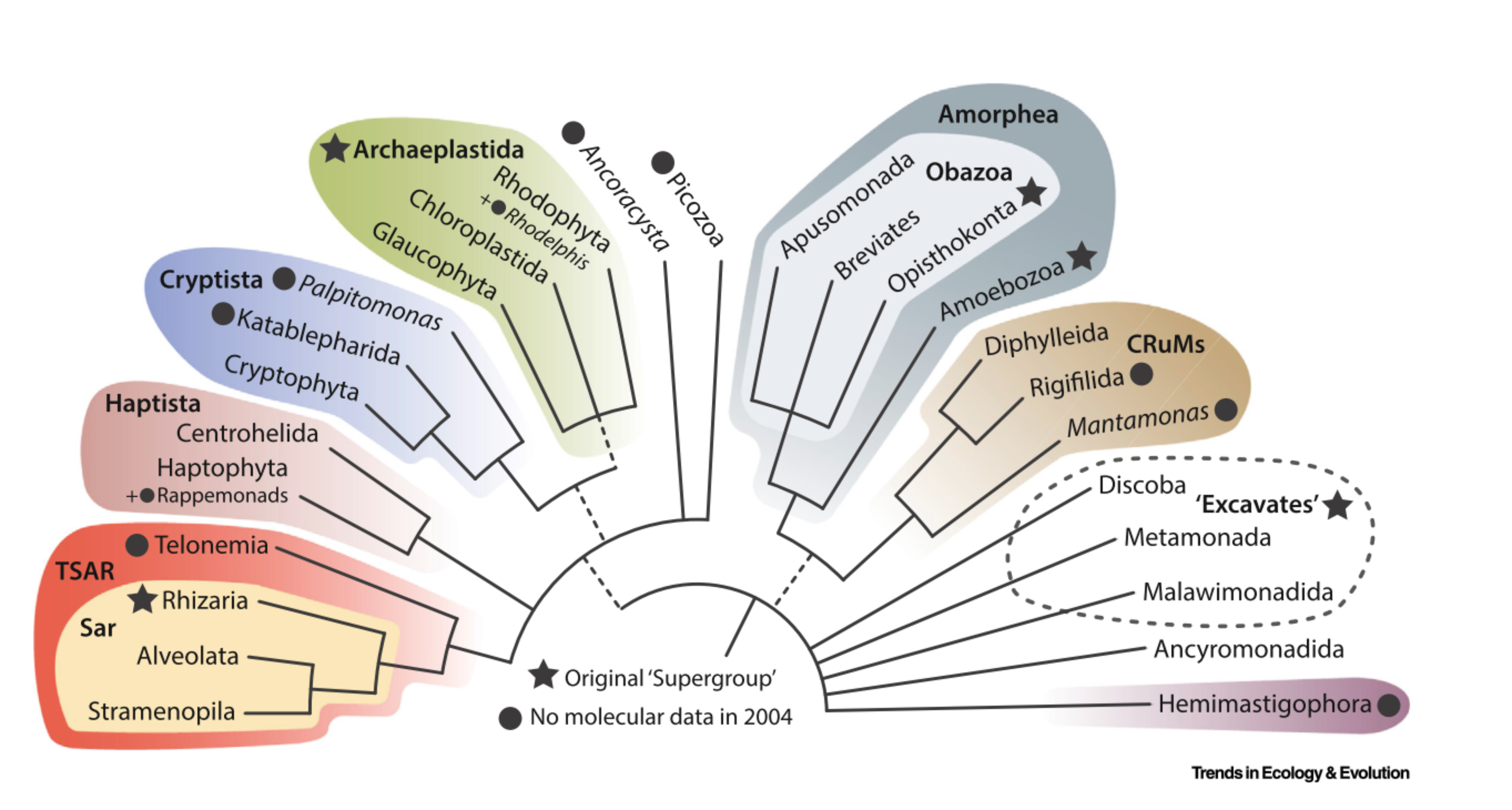
Release 5.0.0 of PR2 (April 2023) included a major update of the taxonomy to include nine taxonomic ranks instead of eight, in order to better accommodate the most recent eukaryote Tree of Life (Burki et al. 2019, Adl et al. 2019). The nine taxonomic ranks now correspond to Domain (replacing Kingdom), Supergroup, Division, Subdivision (new taxonomic rank added), Class, Order, Family, Genus, and Species.
The inclusion of nine taxonomic ranks allows to better match recent phylogenomic evidence, e.g. the new supergroup TSAR in PR2 version 5 now subsumes four supergroups in previous PR2 versions; namely Alveolata, Rhizaria, Stramenopiles, and Telonemia (Strassert et al. 2019). Similarly, the supergroup Obazoa unites Opisthokonta, Apusomonada, and Breviatea (Brown et al. 2013). Having increased taxonomic ranks also allowed updating the taxonomy to be more informative for groups such as Discoba, where Diplonemea, Euglenida, Kinetoplastea, and Symbiontida could be grouped under the subdivision Euglenozoa (Yubuki et al. 2009, Simpson 1997).
Current supergroups
PR2 version 5 includes nine supergroups. These are:
- Amoebozoa
- Archaeplastida
- CRuMs
- Cryptista
- Haptista
- Excavata
- Obazoa
- TSAR
- Provora (new supergroup added; Tikhonenkov et al. 2022)
The kingdoms of previous PR2 versions and where are they now?
- Amoebozoa is still considered as a supergroup in the new version of PR2.
- Archaeplastida is also still considered as a supergroup in PR2 version 5, and now includes Picozoa (Schön et al. 2021).
- Apusozoa has been shown to be polyphyletic, with Mantamonadidea, Collodictyonidea, and Rigifilida belonging to the supergroup CRuMs (Brown et al. 2018), Ancyromonadida representing a deep branching lineage of unknown phylogenetic affiliation, and the remaining lineages in Apusomonada within Obazoa.
- Hacrobia has similarly been shown to be polyphyletic and its members are now split up between Haptista and Cryptista, with the exception of Telonemia which is in TSAR, and Picozoa in Archaeplastida (Yabuki et al. 2014, Cavalier-Smith et al. 2015, Burki et al. 2016, Strassert et al. 2019, Schön et al. 2021).
- Excavata is still considered a supergroup.
- Opisthokonta is now a division within Obazoa.
- Alveolata is now a division within TSAR.
- Protalveolata has been renamed to Colponemidia, a subdivision with Alveolata, TSAR.
- Rhizaria is now a division with TSAR.
- Stramenopiles is also a division within TSAR.
Problems with taxonomy and metazoa and fungi
References
Adl, S. M., Bass, D., Lane, C. E., Lukeš, J., Schoch, C. L., Smirnov, A., … & Zhang, Q. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. Journal of Eukaryotic Microbiology, 66(1), 4-119. https://doi.org/10.1111/jeu.12691
Brown, M. W., Sharpe, S. C., Silberman, J. D., Heiss, A. A., Lang, B. F., Simpson, A. G., & Roger, A. J. (2013). Phylogenomics demonstrates that breviate flagellates are related to opisthokonts and apusomonads. Proceedings of the Royal Society B: Biological Sciences, 280(1769), 20131755. https://doi.org/10.1098/rspb.2013.1755
Brown, M. W., Heiss, A. A., Kamikawa, R., Inagaki, Y., Yabuki, A., Tice, A. K., … & Roger, A. J. (2018). Phylogenomics places orphan protistan lineages in a novel eukaryotic super-group. Genome Biology and Evolution, 10(2), 427-433. https://doi.org/10.1093/gbe/evy014
Burki, F., Kaplan, M., Tikhonenkov, D. V., Zlatogursky, V., Minh, B. Q., Radaykina, L. V., … & Keeling, P. J. (2016). Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proceedings of the Royal Society B: Biological Sciences, 283(1823), 20152802. https://doi.org/10.1098/rspb.2015.2802
Burki, F., Roger, A. J., Brown, M. W., & Simpson, A. G. (2020). The New Tree of Eukaryotes. Trends in Ecology & Evolution, 35(1), 43-55. https://doi.org/10.1016/j.tree.2019.08.008
Cavalier-Smith, T., Chao, E. E., & Lewis, R. (2015). Multiple origins of Heliozoa from flagellate ancestors: new cryptist subphylum Corbihelia, superclass Corbistoma, and monophyly of Haptista, Cryptista, Hacrobia and Chromista. Molecular Phylogenetics and Evolution, 93, 331-362. https://doi.org/10.1016/j.ympev.2015.07.004
Schön, M. E., Zlatogursky, V. V., Singh, R. P., Poirier, C., Wilken, S., Mathur, V., … & Burki, F. (2021). Single cell genomics reveals plastid-lacking Picozoa are close relatives of red algae. Nature Communications, 12(1), 6651. https://doi.org/10.1038/s41467-021-26918-0
Simpson, A. G. (1997). The identity and composition of the Euglenozoa. Archiv für Protistenkunde, 148(3), 318-328. https://doi.org/10.1016/S0003-9365(97)80012-7{.uri}
Strassert, J. F., Jamy, M., Mylnikov, A. P., Tikhonenkov, D. V., & Burki, F. (2019). New phylogenomic analysis of the enigmatic phylum Telonemia further resolves the eukaryote tree of life. Molecular Biology and Evolution, 36(4), 757-765. https://doi.org/10.1093/molbev/msz012
Tikhonenkov, D. V., Mikhailov, K. V., Gawryluk, R. M., Belyaev, A. O., Mathur, V., Karpov, S. A., … & Keeling, P. J. (2022). Microbial predators form a new supergroup of eukaryotes. Nature, 1-6. https://doi.org/10.1038/s41586-022-05511-5
Yabuki, A., Kamikawa, R., Ishikawa, S. A., Kolisko, M., Kim, E., Tanabe, A. S., … & Inagki, Y. (2014). Palpitomonas bilix represents a basal cryptist lineage: insight into the character evolution in Cryptista. Scientific Reports, 4(1), 4641. https://doi.org/10.1038/srep04641
Yubuki, N., Edgcomb, V. P., Bernhard, J. M., & Leander, B. S. (2009). Ultrastructure and molecular phylogeny of Calkinsia aureus: cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiology, 9(1), 1-22. https://doi.org/10.1186/1471-2180-9-16