PR2 version 5.0.0: 9 levels taxonomy and other changes
We are very pleased to announce the new PR2 version 5.0.0.
Very big thanks to all the contributors - see list.
Taxonomy evolving from 8 to 9 levels
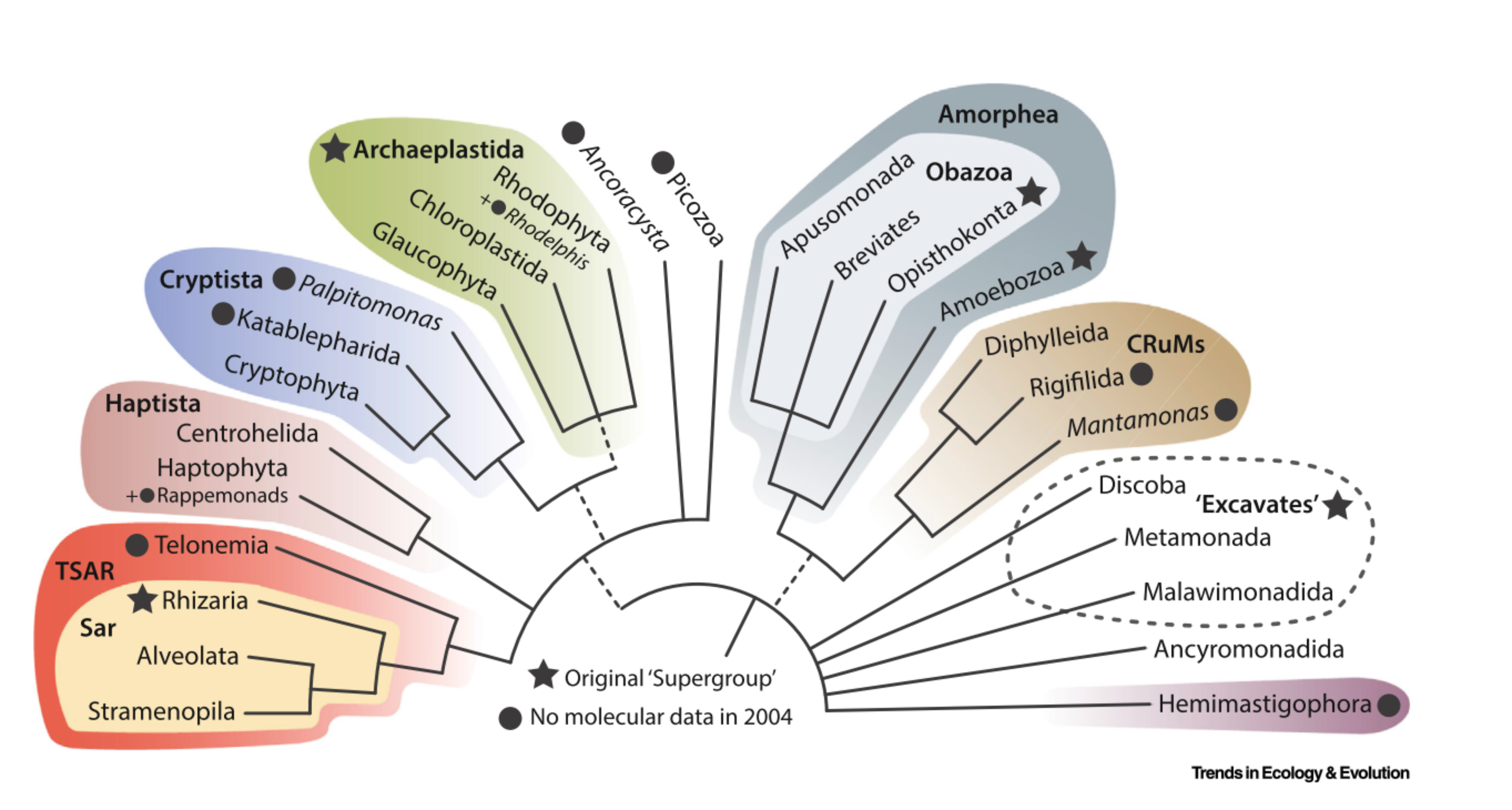
Eukaryote taxonomy is evolving very quickly fostered by the discovery of novel taxa and by phylogenomics. Therefore the structure of taxonomy database needs to evolve to reflect these changes. Following the structure outline by Burki et al. (2019) we modified the primary structure of the PR2 database adding a new level subdivision between division and class. Moreover the top level previously called kingdom has been renamed to domain. These changes and their rationale are detailed in the following article.
Taxonomy updates
The taxonomy of a number of major groups has been updated.
Bacteria and Archaea
- Supergroups have been added.
Stramenopiles
- Diatoms: Classes which were missing from previous versions have been added from WoRMS and Algaebase.
- Chrysophyceae: The taxonomy of the full class has been updated.
- Olisthodiscophyceae : new class added.
Alveolates
- Ciliates: Finally Spirotrichea have been updated to be coherent with the other ciliates groups as annotated in the EukRef workshop of 2016 and new sequences added.
- Dinoflagellates: A number of groups have updated taxonomy and new sequences added.
- Perkinsea: The taxonomy of the group has been completely updated and new sequences added.
Fungi
- Chytrids and Fungi_X: Taxonomy has been completely edited and new sequences added.
Others groups
- New organelles sequences (plastids and mitochondria)
- Spumellaria, Percolomonads, Picozoa, Microsporidia: taxonomy updated and sequences added
- Myxogastria taxonomy updated.
- New supergroup added: Provora
Link to other databases
Although PR2 tries to keep updated annotations, it is necessary to also correlate this information such as Silva or EukRibo, a newly developed SSU rRNA database. Another important link is with functional traits associated to the taxa or to the sequences. Although some of the fields of PR2 provide some functional information, novel database are developed such as the Mixoplankton Database which provides a reference list of planktonic taxa wit mixotrophic capacity. In this new update of PR2 we have linked this information to the PR2 entries. More will come in the future as more functional databases are developed.
Eukribo
EukRibo is a new database that has developed by Berney et al. (2023) and available from Zenodo. It is characterized by a very high quality of annotation. It also labels sequences that contain either the V4 or V9 region.
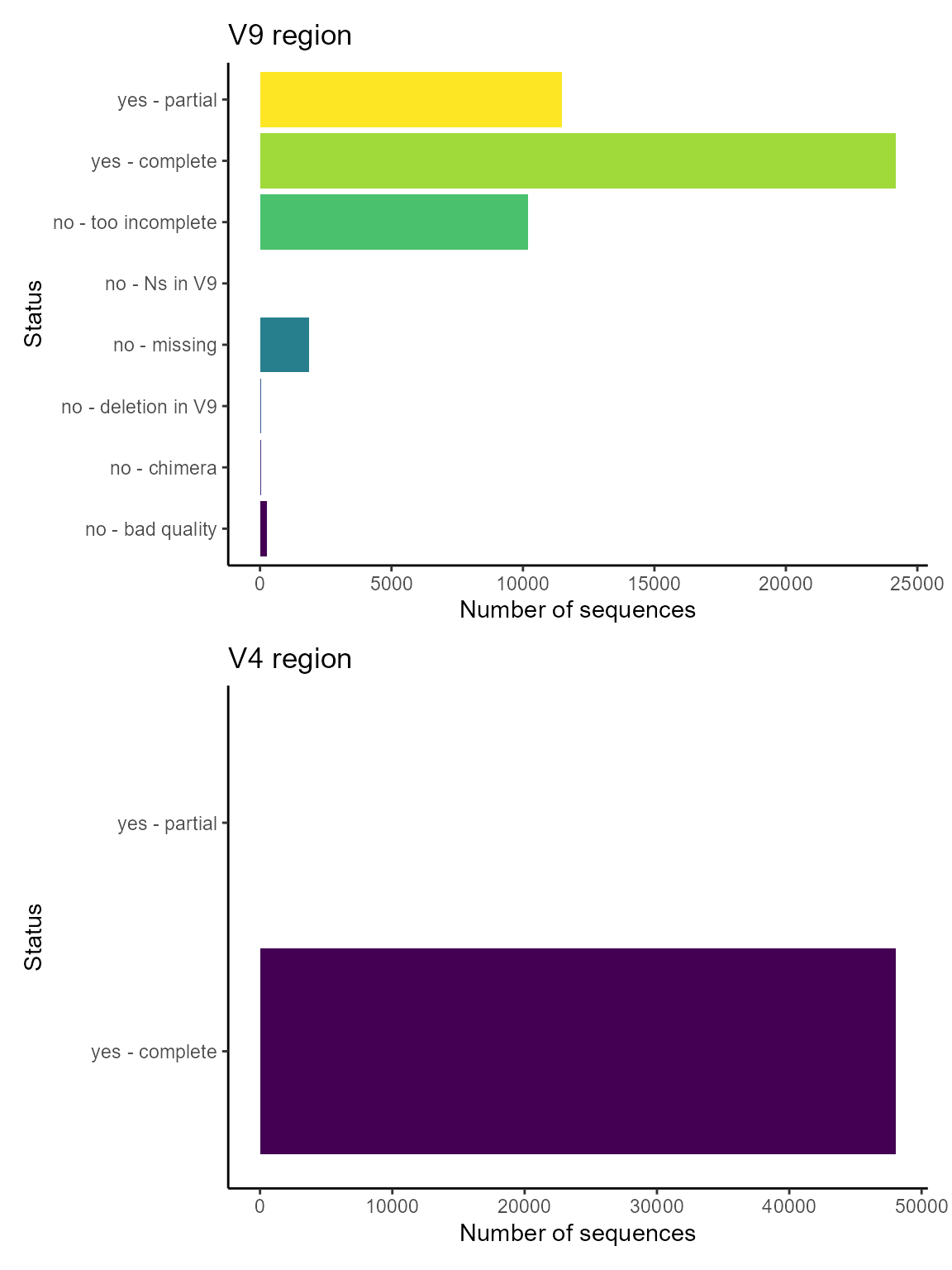
Please see this tutorial to make use of this new PR2 feature.
Mixoplankton Database
The Mixoplankton Database (MDB, DOI 10.5281/zenodo.7560582) has been developed by Mitra et al. 2023 (https://doi.org/10.1111/jeu.12972). It tabulates species that belong to the mixoplankton. This database has been linked to the PR database which contains a new field called mixoplankton:
* CM - Constitutive Mixoplankton
* GNCM - Generalist Non-Constitutive Mixoplankton
* pSNCM - plastidic Specialist Non-Constitutive Mixoplankton
* eSNCM - endosymbiotic Specialist Non-Constitutive Mixoplankton

Please see this tutorial to make use of this new PR2 feature.
WoRMS database
WoRMS database is an authoritative and comprehensive list of names of marine organisms, including information on synonymy. The content of WoRMS is controlled by taxonomic and thematic experts, not by database managers. For species in PR2 that have an entrin WoRMS we have now added a link to the AphaID (worms_id field) as well as information on the distribution of the species (maine, brackish, freshwater, terrestrial).
The complete list of changes is available from here.
Please let us know of any problem you encounter with this release on GitHub.